
Homologous recombination deficiency score decreased after neoadjuvant chemotherapy in ovarian cancer patient: a case report
Highlight box
Key findings
• We reported one high-grade serous ovarian cancer case whose homologous recombination status changed from homologous recombination deficiency (HRD) positive to HRD negative after neoadjuvant chemotherapy (NAC).
What is known and what is new?
• Clinical tests aim to predict the presence of HRD by detecting genomic features, which are considered permanent “genomic scars” and confer sensitivity to poly(ADP-ribose) polymerases inhibitor (PARPi). However, it is still unknown whether the HRD remains unchanged and whether residual tumors respond to PARPi after NAC.
• The phenotypic measurements of HRD might change between original tumors and post-NAC tumors, affecting treatment decisions.
What is the implication, and what should change now?
• Our discovery suggests that extra precautions need to be taken when interpreting HRD score results for patients receiving NAC, especially for BRCA-negative patients. It is important to design future studies that include multi-sampling to allow the assessment of HRD status dynamically.
Introduction
Background
Ovarian cancer is the seventh most commonly diagnosed cancer among women and is the leading cause of death amongst women with gynecological cancers (1). A 5-year survival among patients with advanced disease [International Federation of Gynecology and Obstetrics (FIGO) stage III–IV] is <30% (2). Epithelial ovarian cancer is the most common type of ovarian cancer and has four main histological subtypes: serous, endometrioid, mucinous, and clear cell. Almost 70% of all epithelial tumors are aggressive high-grade serous ovarian cancer (HGSOC) and present in advanced stages (3). Cytoreductive surgery followed by platinum-based chemotherapy has remained the mainstay of treatment for decades (4). Homologous recombination deficiency (HRD) is a functional defect in the homologous recombination (HR) DNA repair pathway. Approximately 50% of HGSOC exhibit HRD, which confers sensitivity to poly(ADP-ribose) polymerases inhibitor (PARPi) and DNA-damaging agents (5). In addition, HRD tumors have been suggested to be more immunogenic, therefore, more susceptible to immunotherapy (6,7).
Rationale and knowledge gap
Clinical tests aim to predict the presence of HRD by detecting genomic features caused by HRD, such as loss of heterozygosity (LOH) (8), telomeric allelic imbalance (TAI) (9), and large-scale state transitions (LSTs) (10). These assays measure the consequence of HRD, irrespective of the underlying etiology. Although genomic alterations induced by HRD are considered permanent “genomic scars”, tumoral HRD status has been recognized to be dynamic over time, such as the acquisition of BRCA reversion mutations that restore HR function in BRCA-mutant cancer cells (11,12). Studies have shown that 25% to 50% of patients treated with PARPi will have relapsed by 30 months from diagnosis, the BRCA reversion may be the underlying mechanism of PARPi resistance (13,14). Neoadjuvant chemotherapy (NAC) is widely applied in routine practice for ovarian cancer. However, it is still unknown whether the phenotypic measurement of HRD remains the same after NAC and whether residual tumors after NAC respond to PARPi based on synthetic lethality.
Objective
Here, we reported one HGSOC case whose HR status changed from HRD positive to HRD negative after NAC. To our knowledge, there is no report documenting the decrease of HRD score in HGSOC patients after NAC. Tumor heterogeneity (intratumoral, intertumoral), and NAC response heterogeneity may explain the HRD status change. This finding indicated that the HR status’s phenotypic measurements of the post-NAC tumor might differ from the original tumor, and extra precautions must be taken when interpreting HRD test results. To make optimal clinical decisions, whether the HRD status of HGSOC patients receiving NAC should be detected from either treatment naïve biopsies, treated tumor samples, or multiple tumor lesions needs thorough study in the future. We present this case in accordance with the CARE reporting checklist (available at https://gpm.amegroups.com/article/view/10.21037/gpm-23-19/rc).
Case presentation
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This research proposal was covered by approval for data use and clinical studies from the General Ethics Commission, West China Second University Hospital. Additional ethical approval was not necessary from the Ethics Commission of West China Second University Hospital, as only anonymized data were analyzed in this study. Written informed consent to participate was given by the patient prior to examination and documentation. A copy of the written consent is available for review by the editorial office of this journal.
The patient was a 65-year-old Chinese woman with no relevant past medical history who presented to hospital on July 2020 with bloating, poor appetite, fatigue, and weight loss (2 kg) for 2 months. Pelvic examination showed healthy external genitalia and cervix. Irregular cystic solid mass could be palpated on both side adnexa. A computed tomography (CT) scan revealed a cystic tumor in the pelvic cavity that involved both side ovaries, with peritoneal carcinomatosis and nodules in the right lobe of the liver. Her cancer antigen 125 (CA-125) level was 584.7 U/mL (normal range of CA-125 level <35 U/mL). A standard laparoscopic procedure was performed for pelvic biopsies under general anesthesia conditions. The endoscopic evaluation of the pelvis showed bloody peritoneal fluid (200 mL), a cystic solid mass involving bilateral adnexa, and extensive peritoneal dissemination. Tissue samples from the cystic solid mass and omental nodules were biopsied for pathologic examination, and ascites were sent for cytology test. The biopsies confirmed high-grade serous adenocarcinoma in the fimbriae end of the fallopian tube, omentum, and ascites (Figure 1). After genetic counseling and informed consent, we assessed her HRD score [PARP inhibitor CDx genetics testing (HRD), BGI, Shenzhen, China] using samples from the biopsy and detected mutations of HR repair (HRR) pathway genes [PARP inhibitor CDx genetics testing (HRR), BGI, Shenzhen, China]. The HRD score comprehensively evaluated LOH, TAI and LST scores with correction of ploidy and purity of tumor by BGI, and the calculation formula of HRD score was preliminarily determined as follows: HRD_{score} = LOH + TAI + (LST − K × Ploidy) (15). The results showed that her HRD score was 42.57 (LOH 21, TAI 10, LST 43, Ploidy 2.03, K 15.5), and no deleterious mutation of any HRR gene was detected either in the germline testing (Table 1) or the somatic testing (Table 2). HRD positive was defined as a high HRD score (above the HRD threshold, ≥30) and/or harmful mutations were detected in tumor BRCA1/2. HRD negative was defined as a low HRD score (below the HRD threshold, <30), and no harmful mutations were detected in tumor BRCA1/2 (15). Based on this, we concluded that her tumor was HRD positive and that she would potentially benefit from PARPi.
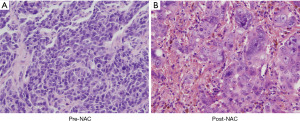
Table 1
| Number | Gene | Transcript | Nucleotide change | Amino acid change | Gene subregion | Heterozygosity | Mutation type | Variant classification |
|---|---|---|---|---|---|---|---|---|
| 1 | ATM | NM_000051.3 | c.1236-3dupT | – | IN9 | Het | Splice | Benign |
| 2 | BARD1 | NM_000465.3 | c.216-14delT | – | IN2 | Het | Splice | Benign |
| 3 | BLM | NM_000057.3 | c.2555+7T>C | – | IN12 | Het | Splice | Benign |
| 4 | BRCA1 | NM_007294.3 | c.4837A>G | p.Ser1613Gly | EX16 | Het | Missense | Benign |
| 5 | BRCA1 | NM_007294.3 | c.3548A>G | p.Lys1183Arg | EX11 | Het | Missense | Benign |
| 6 | BRCA1 | NM_007294.3 | c.3113A>G | p.Glu1038Gly | EX11 | Het | Missense | Benign |
| 7 | BRCA1 | NM_007294.3 | c.2612C>T | p.Pro871Leu | EX11 | Het | Missense | Benign |
| 8 | BRCA2 | NM_000059.3 | c.943T>A | p.Cys315Ser | EX10 | Het | Missense | Likely benign |
| 9 | BRCA2 | NM_000059.3 | c.7806-14T>C | – | IN16 | Hom | Splice | Benign |
| 10 | BRIP1 | NM_032043.2 | c.2755T>C | p.Ser919Pro | EX19 | Hom | Missense | Benign |
| 11 | BRIP1 | NM_032043.2 | c.587A>G | p.Asn196Ser | EX6 | Het | Missense | Likely benign |
| 12 | CDH1 | NM_004360.4 | c.48+6C>T | – | IN1 | Hom | Splice | Benign |
| 13 | CDH1 | NM_004360.4 | c.2164+17dupA | – | IN13 | Het | Splice | Benign |
| 14 | CFTR | NM_000492.3 | c.1408G>A | p.Val470Met | EX11 | Het | Missense | Benign |
| 15 | CHEK1 | NM_001274.5 | c.1411A>G | p.Ile471Val | EX13 | Hom | Missense | Benign |
| 16 | EPCAM | NM_002354.2 | c.344T>C | p.Met115Thr | EX3 | Het | Missense | Benign |
| 17 | ERCC2 | NM_000400.3 | c.2251A>C | p.Lys751Gln | EX23 | Het | Missense | Benign |
| 18 | ERCC2 | NM_000400.3 | c.934G>A | p.Asp312Asn | EX10 | Het | Missense | Benign |
| 19 | ERCC2 | NM_000400.3 | c.477+9A>C | – | IN6 | Het | Splice | Benign |
| 20 | ERCC4 | NM_005236.2 | c.974-7G>A | – | IN5 | Hom | Splice | Benign |
| 21 | ERCC5 | NM_000123.3 | c.3310G>C | p.Asp1104His | EX15 | Hom | Missense | Benign |
| 22 | FANCA | NM_000135.2 | c.3935-16C>T | – | IN39 | Het | Splice | Benign |
| 23 | FANCA | NM_000135.2 | c.2426G>A | p.Gly809Asp | EX26 | Hom | Missense | Benign |
| 24 | FANCA | NM_000135.2 | c.2151+8T>C | – | IN23 | Hom | Splice | Benign |
| 25 | FANCA | NM_000135.2 | c.1826+15T>C | – | IN20 | Hom | Splice | Benign |
| 26 | FANCA | NM_000135.2 | c.1501G>A | p.Gly501Ser | EX16 | Hom | Missense | Benign |
| 27 | FANCA | NM_000135.2 | c.1226-20A>G | – | IN13 | Hom | Splice | Benign |
| 28 | FANCA | NM_000135.2 | c.796A>G | p.Thr266Ala | EX9 | Hom | Missense | Benign |
| 29 | FANCA | NM_000135.2 | c.710-12A>G | – | IN7 | Hom | Splice | Benign |
| 30 | FANCD2 | NM_001018115.2 | c.378-6_378-5delTT | – | IN5 | Het | Splice | Benign |
| 31 | FANCD2 | NM_001018115.2 | c.439-16A>G | – | IN6 | Het | Splice | Benign |
| 32 | FANCD2 | NM_001018115.2 | c.695+16G>C | – | IN9 | Het | Splice | Benign |
| 33 | FANCD2 | NM_001018115.2 | c.784-19C>T | – | IN10 | Het | Splice | Benign |
| 34 | FANCD2 | NM_001018115.2 | c.1278+1delG | – | IN15 | Het | Splice donor | Benign |
| 35 | FANCD2 | NM_001018115.2 | c.1278+3_1278+5delAAG | – | IN15 | Het | Splice | Benign |
| 36 | FANCD2 | NM_001018115.2 | c.1278+15C>T | – | IN15 | Het | Splice | Benign |
| 37 | FANCD2 | NM_001018115.2 | c.3466+34_3466+36dupTTT | – | IN34 | Het | Splice | Benign |
| 38 | FANCD2 | NM_001018115.2 | c.3849+13A>G | – | IN38 | Het | Splice | Benign |
| 39 | FANCI | NM_001113378.1 | c.257C>T | p.Ala86Val | EX4 | Het | Missense | Benign |
| 40 | FANCI | NM_001113378.1 | c.1698+15C>T | – | IN17 | Het | Splice | Benign |
| 41 | FANCI | NM_001113378.1 | c.2225G>C | p.Cys742Ser | EX22 | Het | Missense | Benign |
| 42 | FANCI | NM_001113378.1 | c.3006+15A>C | – | IN27 | Het | Splice | Benign |
| 43 | GEN1 | NM_001130009.2 | c.274T>A | p.Ser92Thr | EX3 | Hom | Missense | Benign |
| 44 | GEN1 | NM_001130009.2 | c.2039C>T | p.Thr680Ile | EX14 | Hom | Missense | Benign |
| 45 | GEN1 | NM_001130009.2 | c.2515_2519delAAGTT | p.Lys839Glufs*2 | EX14 | Het | Frameshift | Benign |
| 46 | MRE11A | NM_005590.3 | c.403-6G>A | – | IN5 | Het | Splice | Benign |
| 47 | MSH2 | NM_000251.2 | c.211+9C>G | – | IN1 | Het | Splice | Benign |
| 48 | MSH2 | NM_000251.2 | c.1661+12G>A | – | IN10 | Het | Splice | Benign |
| 49 | MSH2 | NM_000251.2 | c.2006-6T>C | – | IN12 | Het | Splice | Benign |
| 50 | MSH6 | NM_000179.2 | c.3438+14A>T | – | IN5 | Het | Splice | Benign |
| 51 | MUTYH | NM_001128425.1 | c.1014G>C | p.Gln338His | EX12 | Het | Missense | Benign |
| 52 | NF1 | NM_000267.3 | c.730+32dupT | – | IN7 | Het | Splice | Benign |
| 53 | PMS2 | NM_000535.6 | c.1621A>G | p.Lys541Glu | EX11 | Hom | Missense | Benign |
| 54 | PMS2 | NM_000535.6 | c.1408C>T | p.Pro470Ser | EX11 | Hom | Missense | Benign |
| 55 | PMS2 | NM_000535.6 | c.706-4delT | – | IN6 | Het | Splice | Benign |
| 56 | PMS2 | NM_000535.6 | c.705+17A>G | – | IN6 | Hom | Splice | Benign |
| 57 | POLE | NM_006231.3 | c.6657+16C>T | – | IN47 | Hom | Splice | Benign |
| 58 | POLE | NM_006231.3 | c.6330+15G>A | – | IN45 | Hom | Splice | Benign |
| 59 | POLE | NM_006231.3 | c.3582+17A>G | – | IN29 | Hom | Splice | Benign |
| 60 | PPP2R2A | NM_001177591.1 | c.1095-10A>G | – | IN9 | Het | Splice | Benign |
| 61 | PRSS1 | NM_002769.4 | c.508A>G | p.Lys170Glu | EX4 | Het | Missense | Benign |
| 62 | PTEN | NM_000314.6 | c.802-3dupT | – | IN7 | Het | Splice | Benign |
| 63 | RAD52 | NM_134424.3 | c.186+13A>G | – | IN3 | Het | Splice | Benign |
| 64 | RAD52 | NM_134424.3 | c.-18-20delT | – | IN1 | Het | Splice | Benign |
| 65 | RAD54L | NM_003579.3 | c.408G>C | p.Lys136 | EX6 | Het | Missense | VUS |
| 66 | SLX4 | NM_032444.3 | c.3812C>T | p.Ser1271Phe | EX12 | Het | Missense | Benign |
| 67 | STK11 | NM_000455.4 | c.33G>A | p.Met11Ile | EX1 | Het | Missense | VUS |
| 68 | TP53BP1 | NM_001141980.1 | c.5306-8delT | – | IN24 | Het | Splice | Benign |
| 69 | TP53BP1 | NM_001141980.1 | c.3421A>C | p.Lys1141Gln | EX17 | Het | Missense | Benign |
| 70 | TP53BP1 | NM_001141980.1 | c.1249G>A | p.Gly417Ser | EX11 | Het | Missense | Benign |
| 71 | TP53BP1 | NM_001141980.1 | c.1074C>G | p.Asp358Glu | EX9 | Het | Missense | Benign |
| 72 | WRN | NM_000553.4 | c.1720+15A>G | – | IN14 | Het | Splice | VUS |
| 73 | WRN | NM_000553.4 | c.1982-5delT | – | IN17 | Hom | Splice | Benign |
| 74 | WRN | NM_000553.4 | c.3138+7G>A | – | IN25 | Hom | Splice | Benign |
| 75 | WRN | NM_000553.4 | c.3222G>T | p.Leu1074Phe | EX26 | Hom | Missense | Benign |
| 76 | XRCC3 | NM_001100119.1 | c.722C>T | p.Thr241Met | EX8 | Het | Missense | Benign |
| 77 | XRCC3 | NM_001100119.1 | c.-354-9A>G | – | IN1 | Hom | Splice | Benign |
HRR, homologous recombination repair; Het, heterozygous mutation; Hom, homozygous mutation; VUS, variant of unknown significance.
Table 2
| Sample | Gene | Mutation | Gene subregion | Transcript | Variant frequency (%) | Variation level |
|---|---|---|---|---|---|---|
| Pre-NAC | TP53 | p.Tyr107Serfs*10 (c.320_338delACGGTTTCCGTCTGGGCTT) | EX4 | NM_000546.5 | 31.66 | Tier II |
| Post-NAC | TP53 | p.Tyr107Serfs*10 (c.320_338delACGGTTTCCGTCTGGGCTT) | EX4 | NM_000546.5 | 9.35 | Tier II |
| GEN1 | p.V781L (c.2341G>C) | EX14E | NM_001130009.1 | 10.83 | Tier III |
HRR, homologous recombination repair; NAC, neoadjuvant chemotherapy; Tier II, variants with potential clinical significance; Tier III, variants of unknown clinical significance.
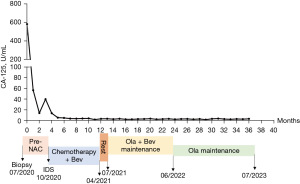
After being evaluated by a gynecological oncologist and calculating the predictive value score using the Multivariate Model of Significant Clinical and Radiologic Criteria Predictive of Suboptimal Cytoreduction system (Suidan score 4, Table 3) (16), the patient was deemed to receive NAC because she was unlikely to achieve residual disease <1 cm only through cytoreduction. Therefore, she was treated with two cycles of carboplatin [area under the curve (AUC) 5] plus paclitaxel (175 mg/m2) (TC, q3w) during July and August 2020, and she had good responses, with the CA-125 level dropping to 14.5 U/mL. Next, the patient received interval debulking surgery (IDS) on October 2020 with the help of a hepatobiliary surgeon (R0 resection), consisting of total hysterectomy with bilateral adnexectomy, pelvic and para-aortic lymph node dissection, omentectomy, appendectomy, and cytoreductive procedure. The postoperative pathology confirmed HGSOC with liver parenchymal metastasis, thus her final diagnosis was stage IVB HGSOC. She recovered well from the surgery and symptoms such as bloating and poor appetite were largely alleviated. She received one cycle of TC (dose same as NAC and frequency), followed by five cycles of TC combined with bevacizumab (7.5 mg/kg) from October 2020 to April 2021. After first-line chemotherapy, her CA-125 level returned to normal (4.0 U/mL), and no tumor activity was seen on the CT scan. According to the National Comprehensive Cancer Network (NCCN) guidelines and the results from the PAOLA-1 trial, which showed improvement in progression-free survival (PFS) when olaparib was added to maintenance bevacizumab in BRCA wild type but HRD positive patients who had a complete response/partial response (CR/PR) after first-line chemotherapy plus bevacizumab (17). The patient received olaparib (150 mg, bid) plus bevacizumab (7.5 mg/kg, q3w) as first-line maintenance therapy from July 2021 to June 2022, followed by single olaparib maintenance with mild adverse events, mainly grade I–II leukopenia and fatigue, without specific interventions. We expect a significant survival benefit from the maintenance therapy due to her HRD positivity. In the most recent follow-up visit (July 2023), her CA-125 was 3.8 U/mL, and no tumor activity was detected on the CT scan (Figure 2). We reassessed the HRD score and HRR pathway genes mutation using the non-necrotic tumor specimen from the IDS after she provided informed consent for testing. No pathogenic mutation of any HRR gene was detected; however, surprisingly, the HRD score calculated from the tumor sample was less than one (LOH 1, TAI 0, LST 0, Ploidy 2.00, K 15.5) and determined as HRD negative. The tumor purity of the tested samples was evaluated by an experienced pathologist, which is 40% pre-NAC and 20% post-NAC. Although tumor purity both fulfilled the quality control requirements for the HRD score assessment, a concern was raised about whether the HRD score change was caused by lower tumor content in the post-NAC sample. To further clarify this issue, we reassessed the HRD score in the post-NAC sample by using tumor materials specifically scraped from tumor sections that an experienced pathologist marked. The results showed that the tumor purity was 50% and an HRD score of −8.93 (LOH 11, TAI 7, LST 5, ploidy 2.06, K 15.5). The average sequencing depths of the three samples are more than 150×, and the capture efficiencies are more than 45% for all samples. These details exclude our concern that the HRD status change observed in this patient was due to lower tumor purity and unqualified quality control.
Table 3
| Criteria | Score |
|---|---|
| Clinical criteria | |
| Age ≥60 years | 1* |
| CA-125 ≥600 U/mL | 1 |
| ASA 3–4 | 3 |
| Radiologic criteria | |
| Retroperitoneal lymph nodes above the renal hilum (including supradiaphragmatic) >1 cm | 1 |
| Diffuse small bowel adhesions/thickening | 1* |
| Perisplenic lesion >1 cm | 2 |
| Small bowel mesentery lesion >1 cm | 2* |
| Root of the superior mesenteric artery lesion >1 cm | 2 |
| Lesser sac lesion >1 cm | 4 |
*, criteria that fit for the patient. Score 0–2: PDS; Score ≥3: NAC + IDS. CA-125, cancer antigen 125; ASA, American Society of Anesthesiologists; PDS, primary debulking surgery; NAC, neoadjuvant chemotherapy; IDS, interval debulking surgery.
Discussion
Key findings
In this study, we reported one HGSOC case whose HR status changed from HRD positive to HRD negative after NAC. This surprising finding indicated that the HR status’s phenotypic measurements might change after NAC, therefore affecting therapeutic decisions.
Strengths and limitations
This report has merits as it is the first report showing the HR status changed from HRD positive to HRD negative after NAC and has educational values. This finding has drawn our attention to the fact that extra precautions need to be taken when interpreting HRD score results for patients receiving NAC. The limitation of our study is that one case was inadequate to draw any statistically meaningful conclusions, and it may lack the ability to generalize.
Comparison with similar researches
To represent, there is no comparable clinical studies reported. Only a previous study (18) reported that concordance in functional HR status between ascites and solid tumor subcultures was seen in only half of the patients which partially supported our theory that intratumoral and intertumoral heterogeneity may be responsible for our discovery.
Explanations of findings
HRD is defined by an impaired error-free HR pathway for repairing DNA double-strand breaks caused by germline/somatic mutations or epigenetic modifications of genes involved in the HR pathway (19). Approximately 50% of HGSOCs exhibit HRD, and the recognition of HRD as a biomarker for HGSOC has transformed treatment paradigms for advanced ovarian cancer. Mechanistically, it is expected that PARPi will be most active in HRD-positive tumors, while HRD-negative tumors are unlikely to respond to PARPi. Although clinical trial results showed that niraparib could provide a longer duration of PFS than placebo in the overall population, niraparib provides a significant clinical benefit for HRD-positive patients over HRD-negative patients (20). Identifying HRD positivity in clinical specimens is critical to patient selection for PARPi, especially for BRCA-negative patients. To date, no uniformly accepted gold standard for HRD assessment exists. Present clinical methods for detecting HRD are limited to direct measuring HR gene mutations, detecting genomic scars reflecting genomic instability, or assessing HR functionality by RAD51 foci formation assays.
Cancer genomes often harbor chromosomal aberrations arising from defective DNA repair, leading to gross chromosomal rearrangements (21) and certain genomic signatures. This leads to the development of assays to evaluate the “genomic scars” as an indirect measure of HRD, such as the presence of LOH, TAI, LST, signature 3 (22), and HRDetect (23). These assays measure the consequence of HRD, irrespective of the underlying etiology. The combination of LOH, TAI and LST performed best at distinguishing HRD-positive tumors from HRD-negative tumors (24,25). More assays are currently commercially available, among them, the ‘myChoice CDx’ assay by Myriad (24), and ‘FoundationFocus CDx BRCA LOH’ by Foundation Medicine (26) have been approved by the Food and Drug Administration (FDA). In this study, the HRD score comprehensively evaluated LOH, TAI, and LST scores with correction of ploidy and purity of tumor by BGI. The HRD score threshold in this study was predefined by analyzing HRD scores in a training cohort of 195 ovarian and breast tumor samples with known BRCA1/2 status and identifying a cutoff with 95% sensitivity to detect those tumors with BRCA1/2 deficiency. HRD positive, defined as an HRD score ≥30 and/or tumor with harmful BRCA1/2 mutation, was tested for its ability to identify which tumors responded to platinum-based chemotherapy in Chinese epithelial ovarian cancer patients (15).
The most likely explanations for HR status change could be intratumoral and intertumoral heterogeneity, and NAC response heterogeneity. First, the HRD score is assessed using core-needle biopsy samples or a few tissue sections from a tumor resection specimen representing only a small fraction of a large tumor mass. Several studies have reported differences in the mutation landscape of the same tumor depending on where the tumor was sampled (27,28). These findings suggest that spatial genomic heterogeneity exists within a single tumor, which might account for the HR status change. Second, tumor cells harboring HRD are more sensitive to platinum-based chemotherapy (29), which results in surviving chemo-resistant clones. These chemo-resistant clones are most probably HRD negative which remained until the surgery and were used for our second test. Last, the tumor tissues analyzed from biopsy and surgery were most probably obtained from different sites of lesions in the same patient. It is likely that biopsy from a single site of lesion may not be representative of the tumor elsewhere genetically and therefore may lead to inconsistent HR status. Recently, single-cell RNA sequencing has been applied to examine how ovarian cancer cells change before and after chemotherapy. No copy number alterations predicted from single-cell RNA sequencing were found, which may imply a stable HR status during chemotherapy. However, direct evidence by measuring HRD on a single cell level is needed (30).
Implications and actions needed
Intertumoral and intratumoral heterogeneity will lead to different prognoses when patients are treated according to the same molecular markers. However, limited by technology and economic costs, tumor molecular tests are usually carried out with samples from a single site. We often test the latest samples in current clinical practice, because previous treatment may affect the tumor genome. This will enable us to consider the follow-up treatments of the patient based on HRD negative status. However, Patel et al. reported that HRD was maintained between the primary and recurrent samples of HGSOC (31). This reminds us that there are still residual tumor cells which are HRD positive, and these cells may predominate at relapse. Thus, we considered treating the patient based on the HRD tested pre-NAC. We will follow up on her treatment and prognosis, which may be presented in the future.
Conclusions
Our discovery suggests that a pre-NAC biopsy sample may be considered for HRD scoring when clinicians encounter the situation we presented here. NAC is widely applied in routine practice, and residual tumors remaining after NAC are believed to be resistant to chemotherapy. However, whether residual tumors after NAC respond to PARPi based on synthetic lethality is still unclear, and this issue needs thorough study in the future. Although repeated tumor testing has not been shown to be of any utility in therapeutic decision-making for patients who have already undergone somatic testing, our finding indicated that it is important to design future studies, including multi-sampling, to allow the assessment of HRD status dynamically.
Acknowledgments
The authors thank Xinyu Yan from BGI Genomics for input and commentary.
Funding: This study was supported by The Key Project of Sichuan Provincial Department of Science and Technology: Study on the key factors affecting the diagnosis and treatment of major diseases in obstetrics and gynecology (19ZDYF, to RY), Science and Technology Department of Sichuan Province, Key R&D Program (2022YFS0080, to QL) and Beijing Kanghua Foundation for the Development of Traditional Chinese and Western Medicine (KH-2021-LLZX-013, to WZ). All funders covered the HRD test cost in this case.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://gpm.amegroups.com/article/view/10.21037/gpm-23-19/rc
Peer Review File: Available at https://gpm.amegroups.com/article/view/10.21037/gpm-23-19/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gpm.amegroups.com/article/view/10.21037/gpm-23-19/coif). R.Y. reported a grant from the Key Project of Sichuan Provincial Department of Science and Technology: Study on the key factors affecting the diagnosis and treatment of major diseases in obstetrics and gynecology (19ZDYF). R.Y. also serves as an unpaid editorial board member of Gynecology and Pelvic Medicine from June 2022 to May 2024. W.Z. reported a grant from Beijing Kanghua Foundation for the Development of Traditional Chinese and Western Medicine (KH-2021-LLZX-013). Q.L. reported grant from Technology Department of Sichuan Province, Key R&D Program (2022YFS0080). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This research proposal was covered by approval for data use and clinical studies from the General Ethics Commission, West China Second University Hospital. Additional ethical approval was not necessary from the Ethics Commission of West China Second University Hospital, as only anonymized data were analyzed in this study. Written informed consent to participate was given by the patient prior to examination and documentation. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Momenimovahed Z, Tiznobaik A, Taheri S, et al. Ovarian cancer in the world: epidemiology and risk factors. Int J Womens Health 2019;11:287-99. [Crossref] [PubMed]
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225-49. [Crossref] [PubMed]
- Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J Clin 2019;69:280-304. [Crossref] [PubMed]
- Piver MS. Treatment of ovarian cancer at the crossroads: 50 years after single-agent melphalan chemotherapy. Oncology (Williston Park) 2006;20:1156, 1158.
- Mirza MR, Coleman RL, González-Martín A, et al. The forefront of ovarian cancer therapy: update on PARP inhibitors. Ann Oncol 2020;31:1148-59. [Crossref] [PubMed]
- van Wilpe S, Tolmeijer SH, Koornstra RHT, et al. Homologous Recombination Repair Deficiency and Implications for Tumor Immunogenicity. Cancers (Basel) 2021;13:2249. [Crossref] [PubMed]
- Bogani G, Lopez S, Mantiero M, et al. Immunotherapy for platinum-resistant ovarian cancer. Gynecol Oncol 2020;158:484-8. [Crossref] [PubMed]
- Abkevich V, Timms KM, Hennessy BT, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 2012;107:1776-82. [Crossref] [PubMed]
- Birkbak NJ, Wang ZC, Kim JY, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2012;2:366-75. [Crossref] [PubMed]
- Popova T, Manié E, Rieunier G, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 2012;72:5454-62. [Crossref] [PubMed]
- Lin KK, Harrell MI, Oza AM, et al. BRCA Reversion Mutations in Circulating Tumor DNA Predict Primary and Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer Discov 2019;9:210-9. [Crossref] [PubMed]
- Weigelt B, Comino-Méndez I, de Bruijn I, et al. Diverse BRCA1 and BRCA2 Reversion Mutations in Circulating Cell-Free DNA of Therapy-Resistant Breast or Ovarian Cancer. Clin Cancer Res 2017;23:6708-20. [Crossref] [PubMed]
- Herzog TJ, Vergote I, Gomella LG, et al. Testing for homologous recombination repair or homologous recombination deficiency for poly (ADP-ribose) polymerase inhibitors: A current perspective. Eur J Cancer 2023;179:136-46. [Crossref] [PubMed]
- Giannini A, Di Dio C, Di Donato V, et al. PARP Inhibitors in Newly Diagnosed and Recurrent Ovarian Cancer. Am J Clin Oncol 2023;46:414-9. [Crossref] [PubMed]
- Chen D, Shao M, Meng P, et al. GSA: an independent development algorithm for calling copy number and detecting homologous recombination deficiency (HRD) from target capture sequencing. BMC Bioinformatics 2021;22:562. [Crossref] [PubMed]
- Suidan RS, Ramirez PT, Sarasohn DM, et al. A multicenter prospective trial evaluating the ability of preoperative computed tomography scan and serum CA-125 to predict suboptimal cytoreduction at primary debulking surgery for advanced ovarian, fallopian tube, and peritoneal cancer. Gynecol Oncol 2014;134:455-61. [Crossref] [PubMed]
- Ray-Coquard I, Pautier P, Pignata S, et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N Engl J Med 2019;381:2416-28. [Crossref] [PubMed]
- O'Donnell RL, Kaufmann A, Woodhouse L, et al. Advanced Ovarian Cancer Displays Functional Intratumor Heterogeneity That Correlates to Ex Vivo Drug Sensitivity. Int J Gynecol Cancer 2016;26:1004-11. [Crossref] [PubMed]
- Hoppe MM, Sundar R, Tan DSP, et al. Biomarkers for Homologous Recombination Deficiency in Cancer. J Natl Cancer Inst 2018;110:704-13. [Crossref] [PubMed]
- González-Martín A, Pothuri B, Vergote I, et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med 2019;381:2391-402. [Crossref] [PubMed]
- Hasty P, Montagna C. Chromosomal Rearrangements in Cancer: Detection and potential causal mechanisms. Mol Cell Oncol 2014;1:e29904. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Davies H, Glodzik D, Morganella S, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017;23:517-25. [Crossref] [PubMed]
- Telli ML, Timms KM, Reid J, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin Cancer Res 2016;22:3764-73. [Crossref] [PubMed]
- Stover EH, Fuh K, Konstantinopoulos PA, et al. Clinical assays for assessment of homologous recombination DNA repair deficiency. Gynecol Oncol 2020;159:887-98. [Crossref] [PubMed]
- Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:1949-61. [Crossref] [PubMed]
- Yates LR, Gerstung M, Knappskog S, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 2015;21:751-9. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Stronach EA, Paul J, Timms KM, et al. Biomarker Assessment of HR Deficiency, Tumor BRCA1/2 Mutations, and CCNE1 Copy Number in Ovarian Cancer: Associations with Clinical Outcome Following Platinum Monotherapy. Mol Cancer Res 2018;16:1103-11. [Crossref] [PubMed]
- Zhang K, Erkan EP, Jamalzadeh S, et al. Longitudinal single-cell RNA-seq analysis reveals stress-promoted chemoresistance in metastatic ovarian cancer. Sci Adv 2022;8:eabm1831.
- Patel JN, Braicu I, Timms KM, et al. Characterisation of homologous recombination deficiency in paired primary and recurrent high-grade serous ovarian cancer. Br J Cancer 2018;119:1060-6. [Crossref] [PubMed]
Cite this article as: Zhang W, Lin X, Yin R, Liao X, Li Q. Homologous recombination deficiency score decreased after neoadjuvant chemotherapy in ovarian cancer patient: a case report. Gynecol Pelvic Med 2023;6:32.


